CURRENT PROJECTS
The Research Alliance Leibniz INFECTIONS continues to focus on antimicrobial resistance (AMR). The World Health Organisation (WHO) describes the spread of AMR as one of the greatest public health threats. A multidisciplinary and interdisciplinary approach is required in order to tackle the problem in a results-oriented and strategically forward-looking manner. Resistant microorganisms not only arise and spread as a result of the selection pressure of antimicrobial therapies, but are also linked to human activities. These include:
- impacts on the environment
- the type of agriculture
- public health
- and the economic and social conditions that determine how medicines are developed, sold and used.
In order to understand why drug resistance develops and how it spreads, experts from many disciplines – not just medicine – need to work closely together. Accordingly, Leibniz INFECTIONS draws on the expertise of 15 Leibniz Institutes and other partners, including the Robert Koch Institute (RKI) and the Friedrich Loeffler Institute (FLI).
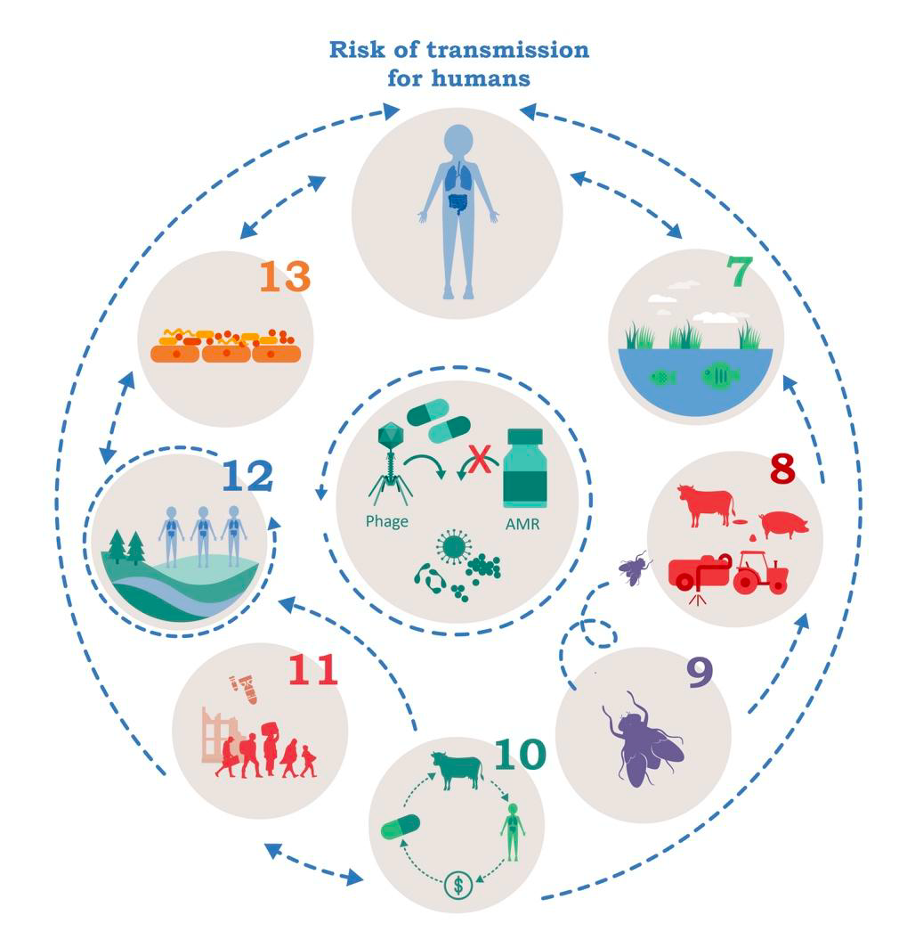
Seven research projects have been designed bringing together various partners. The interdisciplinary project teams (IPTs) are investigating the transmission of AMR from various sources – namely human, environmental, agricultural and socio-economic. The aim is to identify the general drivers of AMR, assess the risk of transmission to human health and develop counter strategies. The IPTs are numbered consecutively from 7 to 13 to distinguish them from IPTs 1 to 6 from the previous funding period (see figure). Further information can be found below in the project descriptions.
Our central objective is to understand the spread of antimicrobial resistance (AMR) between livestock, the environment, and humans. Within the Leibniz Research Alliance INFECTIONS, IPT1 serves as a central hub for bacteriological, molecular biological and spectroscopic identification of AMR bacteria and for bioinformatics analyses of large-scale genomic sequencing data. Metagenomic sequencing of DNA from samples collected within the other IPTs will provide cultivation-independent information on the diversity and abundance of AMR microorganisms. We will combine short-read and long-read DNA sequencing, specific bioinformatics pipelines and public databases to identify antibiotic resistance genes and mutations. Subsequently, specific AMR of interest (e.g. common resistance genes) will be traced across a larger number of samples by using quantitative PCR. Dilution to extinction in selective cultivation media will be used to isolate AMR pathogenic bacteria from environmental samples. Bacterial isolates will be tested for their antibiotic susceptibilities and their cell responses to antibiotics will be characterized by using Raman spectroscopy. Further, the genomes from selected bacterial isolates will be sequenced to track the sources of environmental AMR, such as from flies (IPT4), hygiene studies (IPT5) and open waters (IPT6).
Participating Institutes: DSMZ, IPHT, IGB, IOER, IZW
Antimicrobial resistance (AMR) is becoming an increasingly urgent concern in our world. As more and more microbes develop resistance to antimicrobials, infectious diseases which were commonly able to be treated, become more difficult to deal with. As part of Interdisciplinary Project Team 2 (IPT2) we are primarily focused on microbes which may cause infections of the lung and are well known to develop resistance to antimicrobials. These infections do not usually occur in healthy people, but can occur in those having problems with the immune system or those who suffer from lung conditions, such as cystic fibrosis. Often these microbes do not cause infections alone, but form highly-structured polymicrobial communities, known as “biofilms”. In view of high cell densities and species diversity, it is not surprising that physical and social interactions within mixed biofilms may lead to cooperation or competition between the cells, which in turn may result in remodelling or even evolution of the members of the biofilm community, including the development of antimicrobial resistance to name just one possible outcome of microbial evolution. It is therefore extremely important to understand how the microorganisms evolve and adapt to new environments in mixed biofilm-forming consortia, given the rapid development of antimicrobial resistance and the consequences for the treatment of infectious diseases. Within the framework of the current project we intend to examine the interactions between pulmonary opportunists and arising antimicrobial resistance in mixed biofilms, placing a special focus on bacterial and fungal pathogens affecting cystic fibrosis patients, such as Stenotrophomonas maltophilia and Candida albicans. In order to mimic the conditions of the human respiratory system and achieve the complexity of the in vivo situation in an in vitro model, we will investigate the biofilm communities in physiologically relevant cell culture systems at the air-liquid interface.
Participating Institutes: FZB, HKI, DPZ, DSMZ, LIV, IPHT, ISAS
The rising use of antibiotics in health and agriculture are driving the increase in antimicrobial resistance (AMR). The increasing use of antibiotics in the health- and livestock sectors along with ineffective stewardship have already led to alarming rates of antimicrobial resistant pathogens – a situation with severe consequences for society. Experts estimate that by 2050 ten million people will die each year from common infections that will then be no longer treatable. The central objective of this research project is to support optimal patterns of antibiotic use worldwide. The focus of this project is on two areas in particular: One, reducing the excessive use of antibiotics and two, promoting equal access to quality antibiotics. The excessive use of antibiotics stems from two sources: A lack of stewardship, standards and controls of medicines, and a lack of diagnostic tools that allow distinguishing between bacterial and viral infections – an information essential for treatment that is effective. Hence, this project will assess the role of better diagnostic tools, more information and transparency for curbing antimicrobial resistance. It will assess the cost-effectiveness of a variety of measures and interventions at the micro- and the macro level and provide concrete policy recommendations.
Participating Institutes: BNITM, GIGA, FZB, IfW, RWI
The project focuses on vector ecology. Vectors like filth flies are able to transport and disseminate various pathogens including bacteria, fungi, viruses and parasites. The common house fly (Musca domestica) can transport bacteria over distances of 5-7 km. It breeds in and feeds on decaying organic matter and animal feces. Due to this coprophagic behaviour bacteria may be ingested and can later be transmitted onto human food by regurgitation and defecation. The frequent preventive use of antibiotics on farms and in lifestock production leads to the emergence of antimicrobial resistant bacteria. These can be disseminated by flies from farm environments to urbanized areas causing severe nosocomial infections among citizens. To understand the potential of flies to spread bacteria over different landscape types and from farm environments to urbanized areas, a mark-release-recapture experiment will be conducted. For this purpose, adults of M. domestica are captured, marked by different colours with a permanent dye and released again. The recapture rate will be determined using specialized dipteran traps. The results will help understand the flight range and the habitat binding of the muscid. Strategies to prevent and inhibit the spread of antimicrobial-resistant bacteria by flies will be elucidated. In a further step, the persistence and multiplication of these bacteria in the flies will be investigated under a laboratory setting. The three larval stages of the muscid fly develop in manure and animal feces where they may ingest antimicrobial resistant bacteria excreted by the lifestock animals. Of special intrest is if the ingested bacteria survive the metamorphosis. This indicates that they can remain in the puparium after histolysis or persist in the newly emerged imagos. Altogether, this project will help understand how antimicrobial resistant bacteria survive and persist during the life cycle of flies and how adult flies transport and disseminate them to urbanized areas.
Participating Institutes: ZALF, DSMZ, ATB, IOER
Within Interdisciplinary Project Team 5 (IPT5) the ATB and partners work intensively together to determine the mechanisms of antimicrobial resistance (AMR) transmission in animal husbandry and to evaluate possible intervention measures. We compare AMR occurrence in typical conventionally grown pigs in a standard and improved hygienic environment (by using insecticides, flytraps, increased disinfection and dusting). In addition, the antibacterial activities of different feed additives will be compared to the basic diet based on microbial colonization in the gut of piglets during the early fattening period. In close cooperation with other INFECTIONS partners, the survival and propagation of AMR microbes in pig faeces, dust and flies under all conditions will be analyzed. The abundance of AMR based on bacteriological methods, PCR and cultivation-independent DNA sequencing will be conducted. With support from other IPTs, the findings will guide interventions and potential mitigation strategies to minimize AMR spread in commercial animal husbandry to reduce potential contamination of the environment (e.g. surface water, when the slurry is used as organic fertilizer on agricultural fields).
Participating Institutes: ATB, DSMZ, ZALF, HKI, IPHT, IOER, VRC2
Antimicrobial resistance (AMR) has been declared by the World Health Organization (WHO) as one of the top 10 global public health threats in the presence and near future. According to WHO, AMR will have a disastrous impact within a generation unless an urgent and effective global mitigation plan is finally implemented. In a recent study, data from 204 countries suggest a burden of 1,27 million deaths associated with AMR bacteria in 2019. One of the challenges of monitoring AMR organisms and antimicrobial resistance genes (ARGs) is their ubiquitous distribution, i.e. they are generally found in humans, animals, plants, microbes and the environment like water, soil and air. In particular, water plays a key role as a vector and reservoir for AMR transmission across urban and rural areas. Our preliminary results suggest that the abundance of AMR in urban waters and sediments is substantially higher compared to rural water bodies. In addition, rural lakes sediments and water derived from farmlands is observed to be a major source of environmental AMR due to frequent antibiotics use for cattle raising. The approach defined for Interdisciplinary Project Team 6 (IPT6) seeks to provide further understanding on AMR dispersion from WWTPs to the environment as well as to characterize the humanization of urban water microbiomes. In particular, the use of long-read sequencing in this project will allow to recover mobile genetic elements (MGEs) such as plasmids and genomic islands (GIs) and thus a higher resolution in the metagenome assembly. Also, risk assessment will be performed in order to evaluate the environmental exposure to ARGs. The results from this project will allow to directly link frequency and diversity of AMR and the carrying bacteria to other findings and team up with institutions of the Leibniz Research Alliance (LRA) INFECTIONS, providing a broad overview of AMR profiles in different waters and potential disease vectors of concern in the area of Berlin-Brandenburg.
Participating Institutes: IZW, IGB, LIV, DPZ, GIGA, IOER
Water is vital for human life, but also plays a key role as a vector and reservoir for AMR transmission across urban and rural areas. Our results suggest that AMR abundance and diversity is higher in urban waters and sediments compared to rural water bodies. Wastewater treatment plants (WWTPs) are a major source of AMR in urban water sources. In addition, rural lakes sediments and water derived from farmlands are major sources of environmental AMR.
We will assess the risk of human contact with AMR through environmental water by leveraging our accumulated data from INFECTIONS in the previous funding period and performing bioinformatic comparison with sequences of clinical isolates in our collections. Analyses will be extended to fungal pathogens, including mobile genetic elements. Future water collection will comprise waste water sampled in partner countries where antibiotics / antifungals are used in agriculture. The phage content of the samples will be determined. We are investigating the phageome (all bacteriophages in the water) and whether they might provide promising alternatives to control AMR carrying bacteria. In pilot studies, we will attempt to remove AMR containing bacteria from wastewater using environmentally detected phage in laboratory based studies.
Participating Institutes: ATB, BNITM, DSMZ, FZB, HKI, IGB, IZW
With steadily rising of global meat consumption and the growing number of livestock animals, antibiotic resistances (AMR) spread by livestock manure is an increasingly problem that affects animals, humans, and environmental health. Because manure is used as fertilizer for agricultural fields in the sense of a sustainable circular bioeconomy, there is a potential source of AMR transmission into the human food chain. In the interdisciplinary project team 8 (IPT8), we focus on a One Health approach to track the dynamics of microbial communities and fly larvae in livestock slurry tanks used for agricultural field fertilization. Here, different farm and laboratory slurry storage conditions will be studied with a focus on the persistence of AMR bacteria including foodborne pathogens such as Salmonella, ESBL-producing E. coli and non-tuberculous mycobacteria. We aim to monitor the abundance of AMR and microbiome changes in farm and laboratory slurry tanks. The influence of seasonal temperature changes, slurry type and tank filling levels will be investigated. We will detect and quantify AMR pathogens in slurry with microbiological cultivation. To assess the microbiome diversity in slurry, 16S rDNA sequencing will be performed. The occurrence and uptake of AMR bacteria from slurry by flies will be investigated. The pathogen load of flies and their larvae will be determined by bacteriological and molecular techniques. The results from this research will enable us to deduce practical and effective measures to reduce AMR bacteria in livestock manure by identifying the optimal manure management conditions.
Participating Institutes: ATB, BNITM, DSMZ, FZB, IGB, IZW, ZALF
The interdisciplinary Project Team 9 (IPT9) investigates how flies can spread antimicrobial-resistant (AMR) pathogens from livestock to humans and assesses the resulting health risks. The study focuses on the common housefly (Musca domestica), but also includes other fly species found near livestock. We will conduct field experiments to determine fly species, population densities, movement patterns, and pathogen loads in both Germany and Ghana. Laboratory studies will examine how flies ingest, retain, and shed AMR pathogens like multidrug-resistant enterobacteria, vancomycin-resistant enterococci, non-tuberculous mycobacteria, and Candida species. Using ecological data and experimental findings, models will be developed to predict fly-driven pathogen spread and transmission and evaluate the risk to humans. The team will also assess how environmental factors affect transmission dynamics and test the effectiveness of various control strategies. Multiple Leibniz institutes contribute their expertise in entomology, microbiology, ecology, and risk analysis to deliver a comprehensive understanding of fly-mediated AMR spread.
Participating Institutes: ATB, BNITM, DSMZ, FLI, FZB, HKI, IOER, ZALF
The unregulated and on a massive scale use of antibiotics in livestock farming is a major contributor to the global spread of antimicrobial resistance (AMR). In many low-income settings, lack of veterinary oversight and weak regulatory systems result in inappropriate antibiotic use. IPT10 aims to develop and test scalable interventions to promote responsible antibiotic use in resource-constrained environments.
We combine three strategies: (1) behavior change communication campaigns targeting livestock keepers, (2) the use of affordable molecular diagnostic tools for animal health, and (3) digital platforms for monitoring antibiotic usage and resistance data. The project is implemented in collaboration with partners in Burundi, Rwanda, Tanzania, Ghana, and the Democratic Republic of Congo.
IPT10 builds on ongoing projects: AMRAfrica (a digital monitoring tool), ADA (novel diagnostics), and mobile AMR laboratories operated by the East African Community. These initiatives provide the foundation for piloting and evaluating integrated AMR control strategies. The evidence will support policy development and guide effective stewardship practices in settings where veterinary services are limited.
Participating Institutes: ATB, BNITM, IPHT, RWI
Participating Institutes: BNITM, FZB, IfW, RKI, RWI
Non-tuberculous mycobacteria (NTM) are a diverse group of bacteria commonly found in the environment, particularly in water and soil. They are increasingly causing infections in humans, particularly in individuals with weak immune systems or pre-existing lung conditions such as cystic fibrosis, and are also known to infect a variety of animal species. These infections are often difficult to treat because many NTM are naturally resistant to multiple antibiotics.
It is thought that people get infected with NTM mainly through contact with environmental sources that contain these bacteria. However, which environmental sources currently pose the highest risk to susceptible individuals remains unknown.
This project aims to collect and analyze NTM from different environmental sources like soil and water, as well as from plant and animals across Germany. The bacteria will be examined to determine how common they are, how genetically diverse they are and whether they are resistant to the antibiotics that are normally used to treat NTM disease. These environmental NTM will also be compared to clinical isolates from human patients using modern molecular tools.
The results will help us better understand potential sources and transmission routes of NTM, evaluate infection risks, and contribute to the development of new strategies for prevention and treatment. Promising new therapies, including bacteriophages and novel compounds, will be tested on these strains in collaboration with project IPT13.
Participating Institutes: ATB, DSMZ, FZB, FLI, IGB, IZW, NTUS, RKI, UoS
In IPT13, the complex interactions of polymicrobial species will be investigated further. Once again, lung epithelial air-liquid-interface cells will be used as the model surface to culture the biofilm. As the next stage lung organoids will be established to provide a more complex growth system. Three groups of microorganisms will be studied: bacteria – M. abscessus, S. maltophilia, fungi: C. albicans, C. auris, A. fumigatus, and virus: influenza and SARS-CoV.
The aims of this project will be to assess the interactions of these microbial species by live cell microscopy, analyse how each species responds to each other in a mixed biofilm via proteomics, and explore phage isolates and novel antibiotics against the polymicrobial biofilms for therapeutic control.

